Peanut: Shape Model of Multiple Domains with Shared Boundaries
What is the Use Case?
The peanut_shared_boundary use case demonstrates using ShapeWorks tools to perform shape modelings for anatomies with multiple structures (domains), e.g., joints, with shared boundaries to capture inter-domains correlations and interactions. It also demonstrates the shared mesh boundary and contour extraction tools and runs multi-domain (meshes and contours) optimization on the peanut dataset.
The peanut dataset,consists of two aligned spheres, but one of the spheres is subtracted from the other. There exists a shared surface between the two spheres. The radii of the two spheres vary inversely, ie: as one gets bigger the other gets smaller.
Grooming Steps
This is how the meshes in the dataset look before grooming.

- Remeshing: Meshes are remeshed to ensure uniform vertices.
- Extract Shared Boundary: In this step, we ingest the two original shapes and output three new shapes, two of which correspond to the original shapes and one for the shared boundary.
- Smoothing:Applied laplacian smoothing.
- Extract Contour: The boundary loop of the shared surface is obtained.
Groomed dataset.

Relevant Arguments
--use_subsample --num_subsample --tiny_test
Optimization Parameters
The python code for the use case calls the optimize command of ShapeWorks which reads the project sheet with the shape filenames and optimization parameter values. See Project excel file for details regarding creating the project sheet.
Below are the default optimization parameters for this use case.
# Create a dictionary for all the parameters required by optimization
parameter_dictionary = {
"number_of_particles": 128,
"use_normals": 0,
"normals_strength": 10.0,
"checkpointing_interval": 1000,
"keep_checkpoints": 0,
"iterations_per_split": 1000,
"optimization_iterations": 1000,
"starting_regularization": 10,
"ending_regularization": 1,
"recompute_regularization_interval": 1,
"domains_per_shape": 1,
"relative_weighting": 1,
"initial_relative_weighting": 0.05,
"procrustes_interval": 0,
"procrustes_scaling": 0,
"save_init_splits": 0,
"verbosity": 0
}
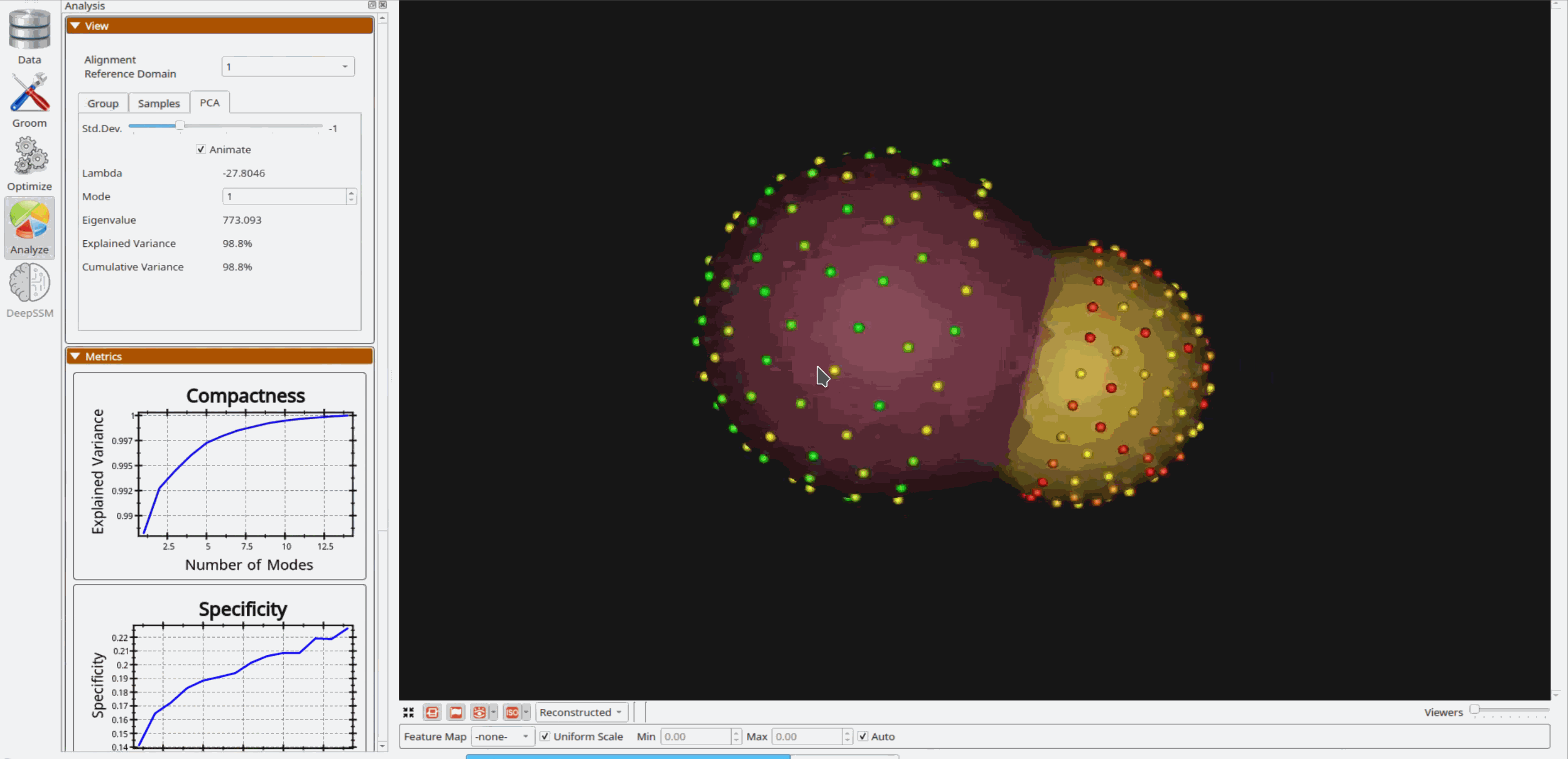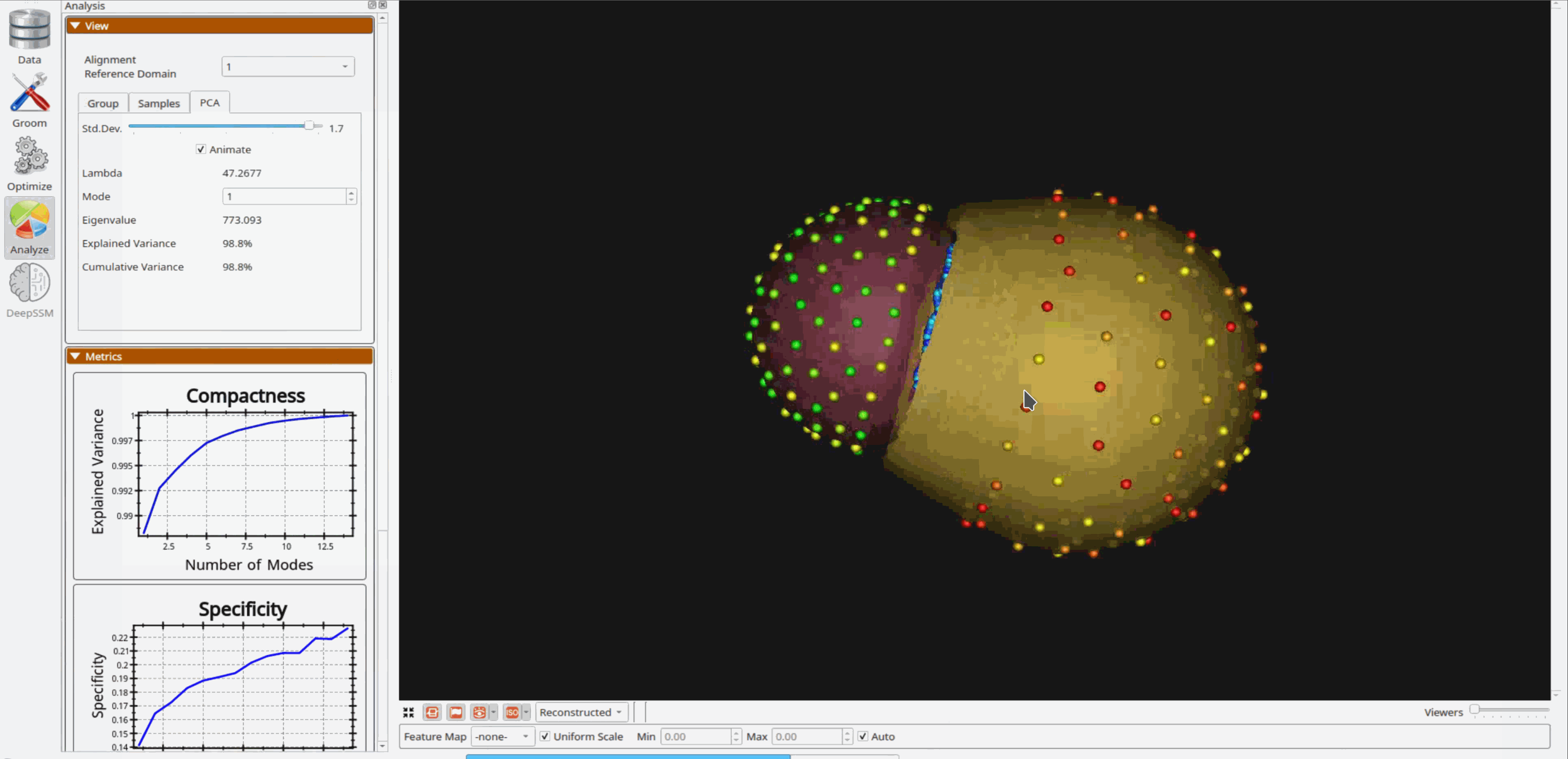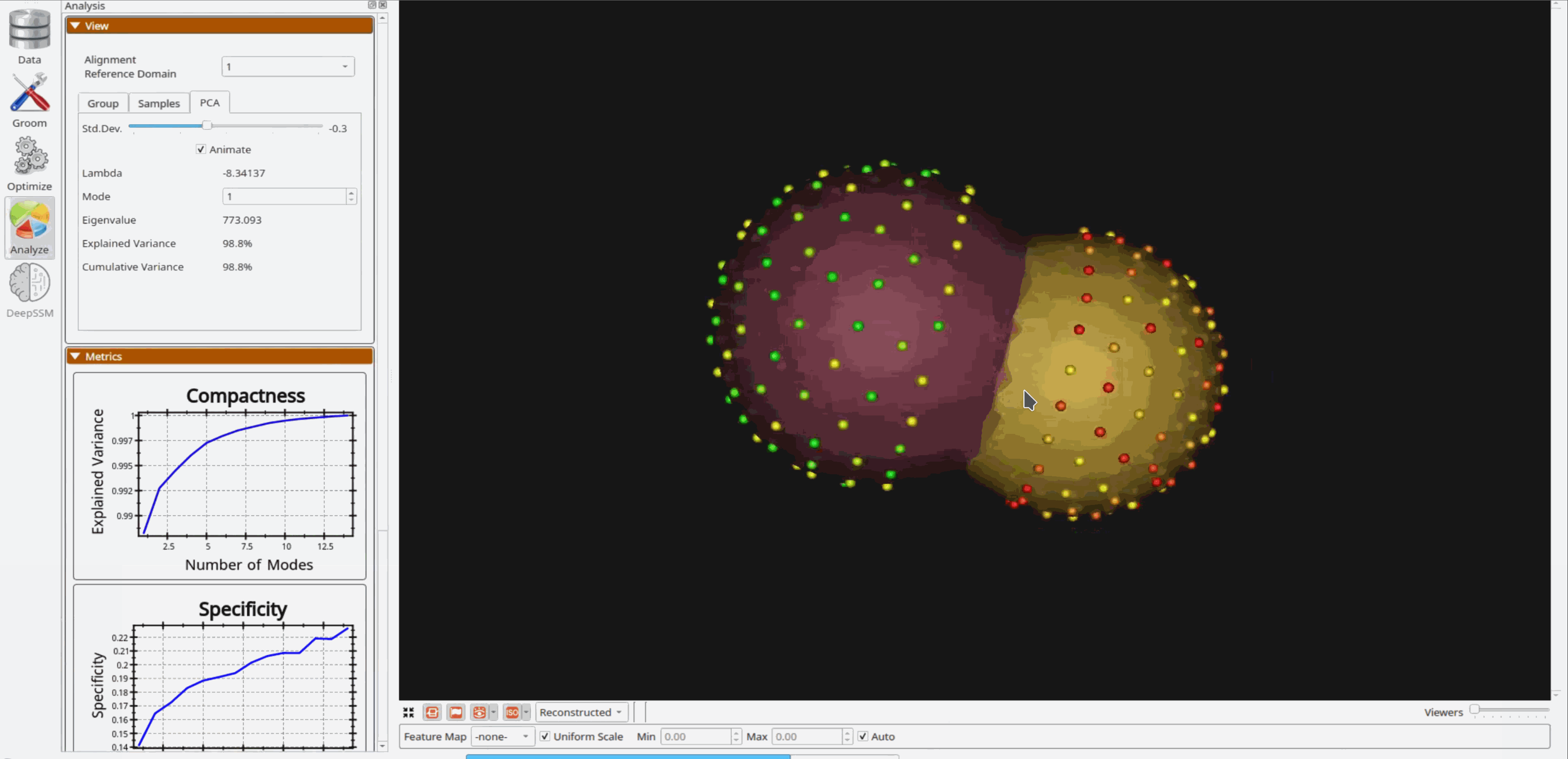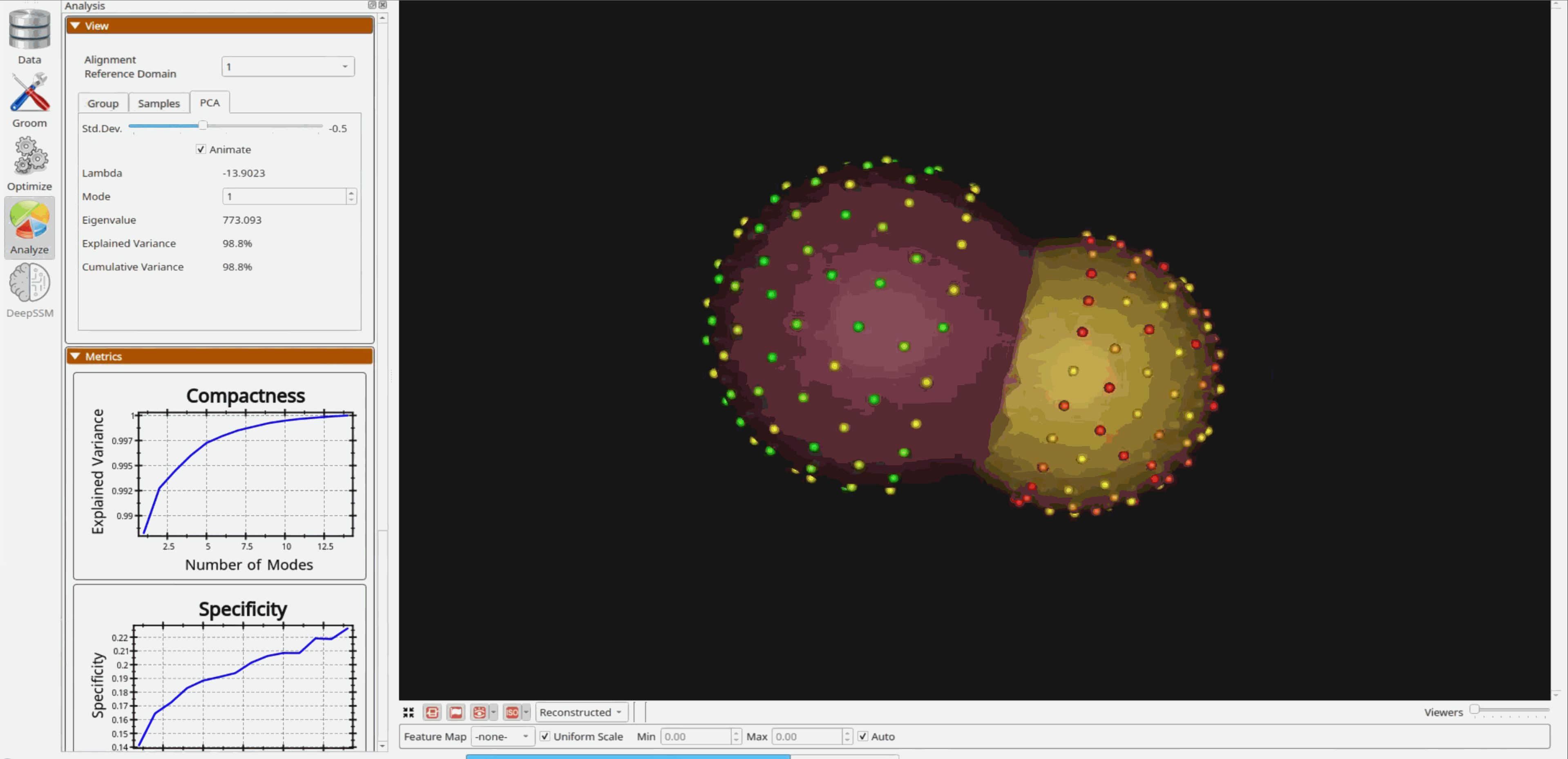
Analyzing Shape Model
Once the python code runs the optimization, the files are saved in the Output folder after which ShapeWorks Studio is launched from the python code to analyze the model.