Shape Model for Multiple Domains directly from Mesh
What is the Use Case?
The hip_multiple_domain use case demonstrates using ShapeWorks tools to perform shape modelings for anatomies with multiple structures (domains). The use case uses the hip joint to demonstrate the capability of ShapeWorks to capture inter-domains correlations and interactions directly on triangular surface meshes. The use case showcases how to calculate the different alignment options available for multiple organ anatomies.
The dataset available for this use case:
- hip - a collection of femur and pelvis bones.
Grooming Data
This is how the meshes in the dataset look before grooming. 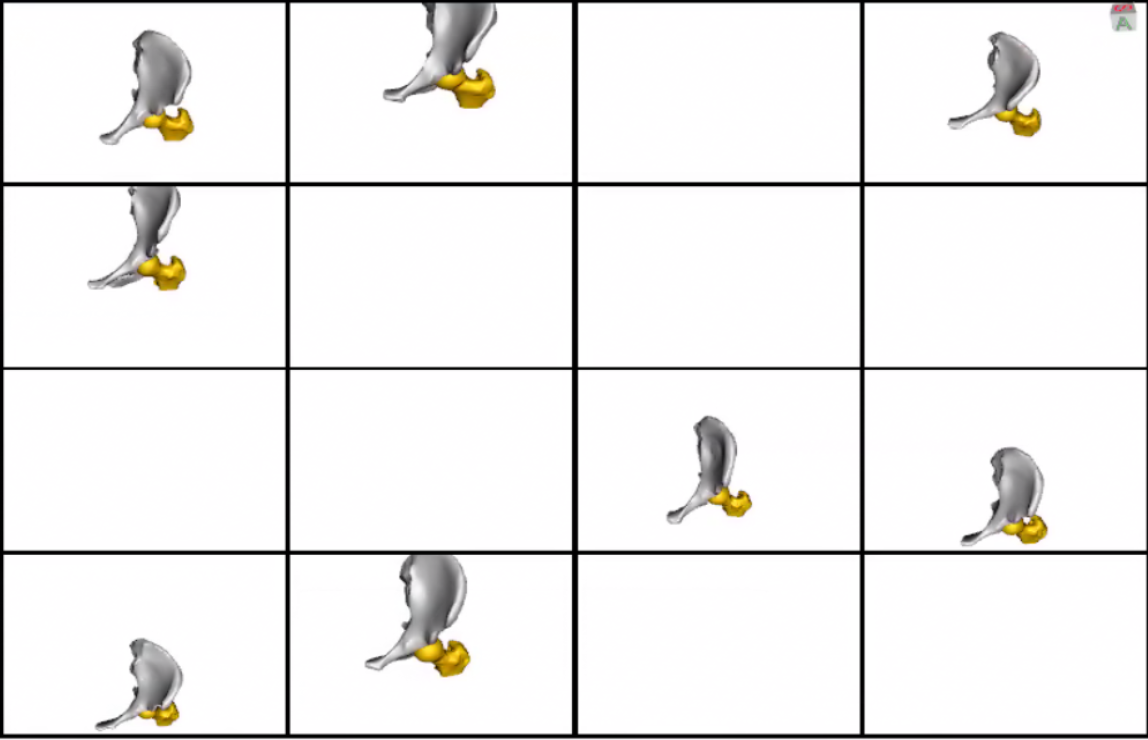
- Smoothing and Remeshing: Meshes are smoothed and remeshed to ensure uniform vertices.
- Reference Selection: The reference is selected by first computing the mean (average) mesh, then selecting the mesh closest to that mean (i.e., medoid).
- Rigid Alignment: This use case calculates three types of alignment: a. Calculate the local alignment matrix using only femurs w.r.t to the femur of the reference shape b. Calcualte the local alignment matrix using only pelvis w.r.t to the pelvis of the reference shape c. Calcualte the global alignment matrix for the complete joint using the reference shape
Here we show how the shapes would look like if the transforms are applied.
Meshes obtained after grooming w.r.t femurs: 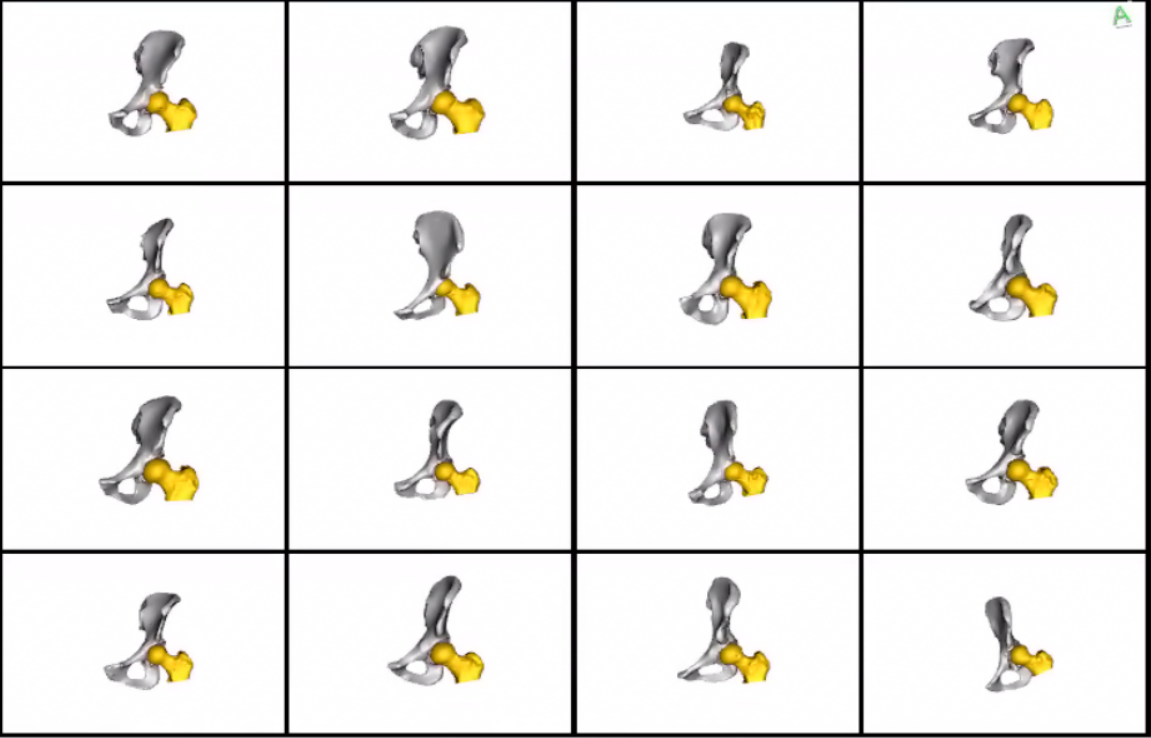 Meshes obtained after grooming w.r.t pelvis:
Meshes obtained after grooming w.r.t pelvis: 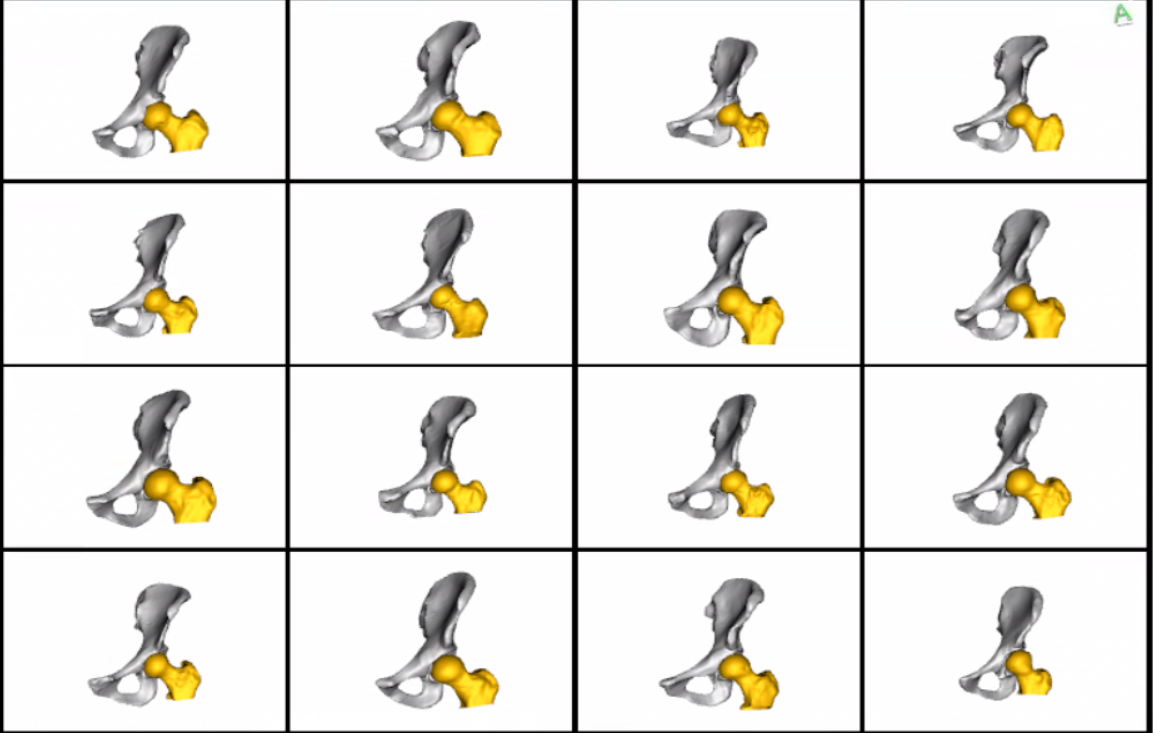 Meshes obtained after grooming w.r.t the whole joint:
Meshes obtained after grooming w.r.t the whole joint: 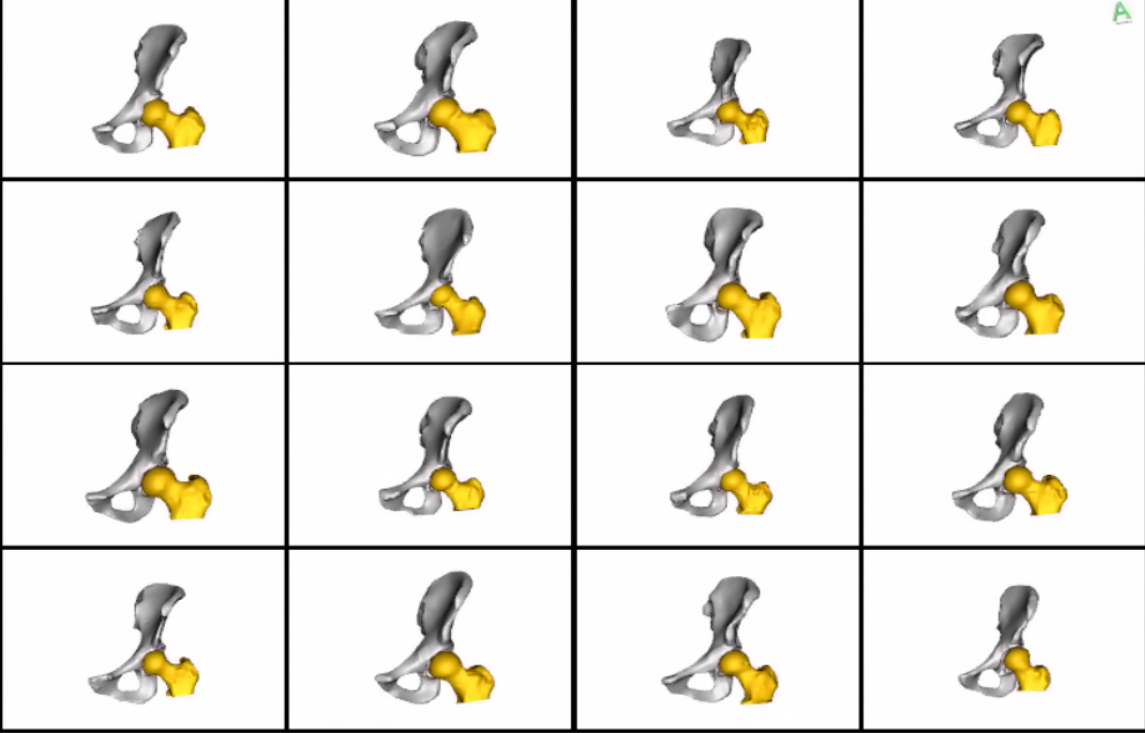
Relevant Arguments
--use_subsample --num_subsample --use_single_scale --tiny_test
Optimization Parameters
The python code for the use case calls the optimize command of ShapeWorks which reads the project sheet with the shape filenames and optimization parameter values. See Optimization parameters for details regarding creating the project sheet.
Below are the default optimization parameters for this use case.
{
"checkpointing_interval" : 200,
"keep_checkpoints" : 0,
"iterations_per_split" : 2500,
"optimization_iterations" : 200,
"starting_regularization" : 1000,
"ending_regularization" : 10,
"relative_weighting" : 3,
"initial_relative_weighting" : 0.05,
"save_init_splits" : 0,
"verbosity" : 0,
"use_normals": 1,
"normals_strength": 5.0,
"procrustes" : 1,
"procrustes_scaling" : 1,
"procrustes_rotation_translation" : 1
}
num_particles = [256,256]
Analyzing Shape Model
Once the python code runs the optimization, the files are saved in the Output folder after which ShapeWorks Studio is launched from the python code to analyze the model.
Here is the optimized shape.
Modes of variation of the hip joint with global alignment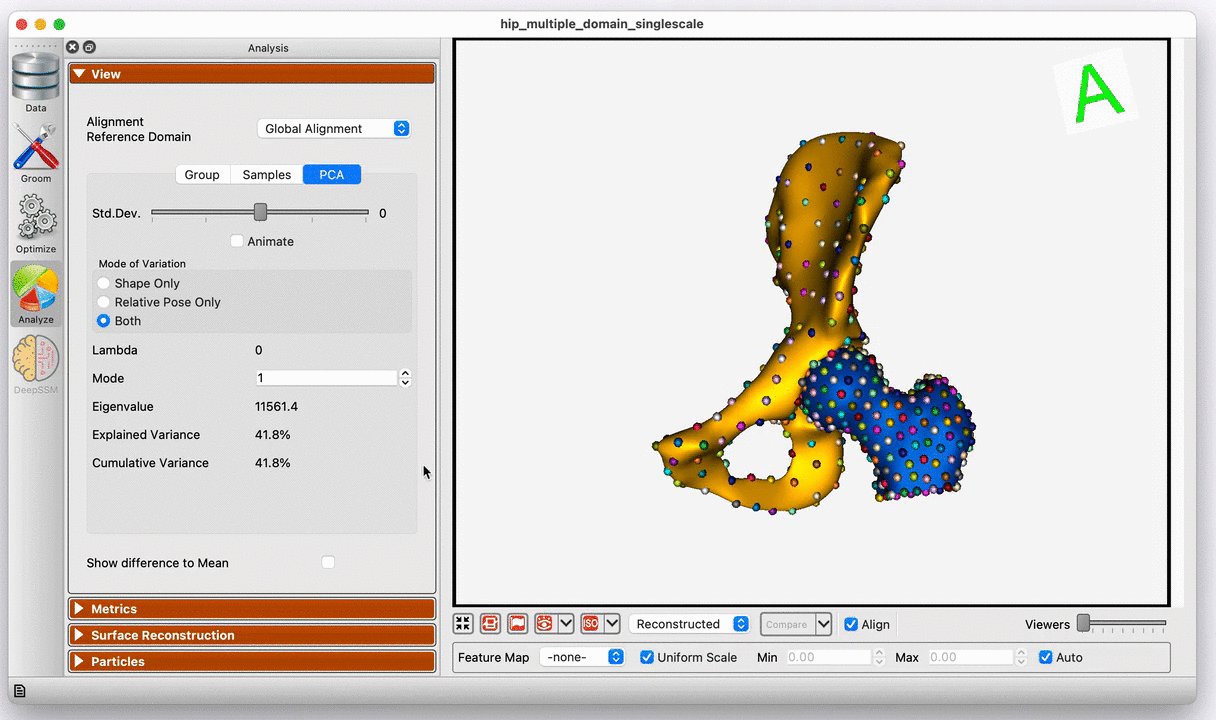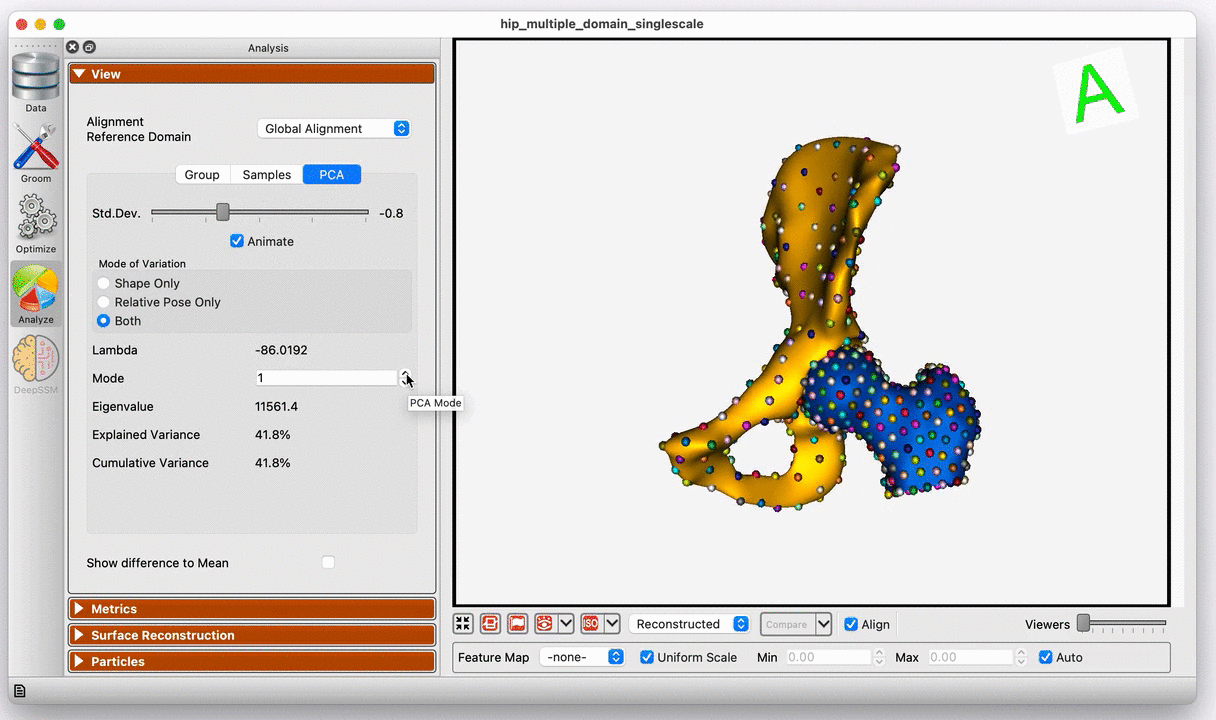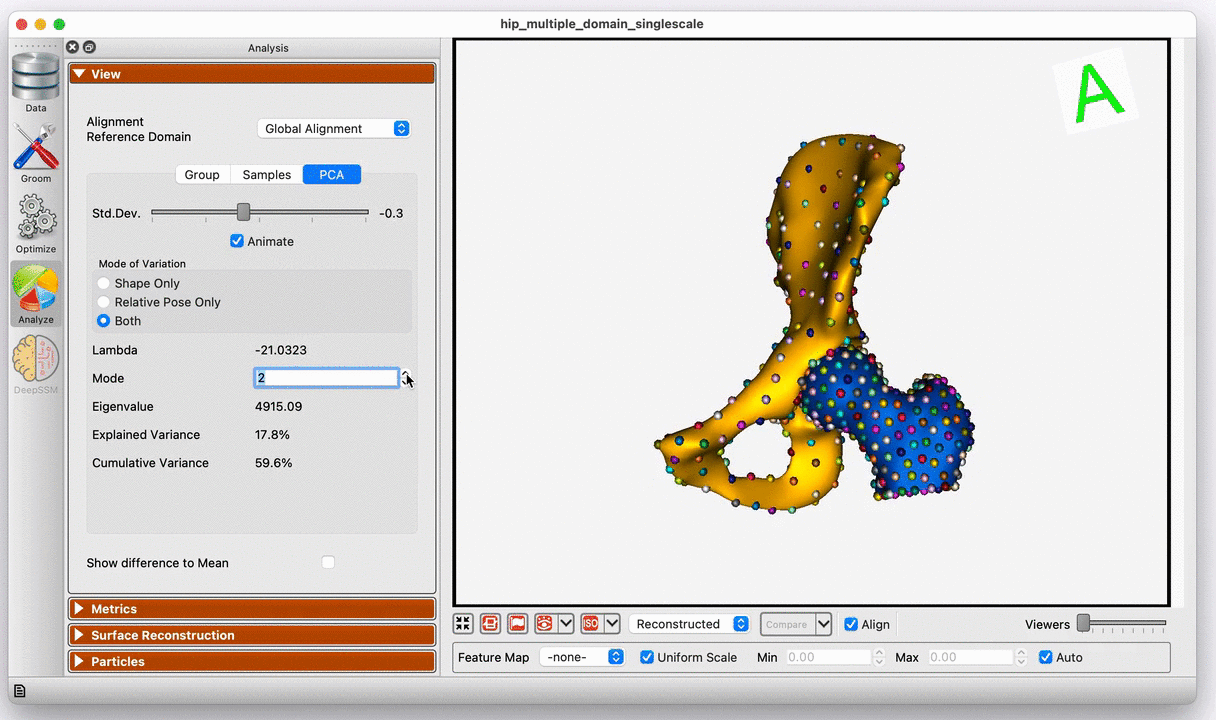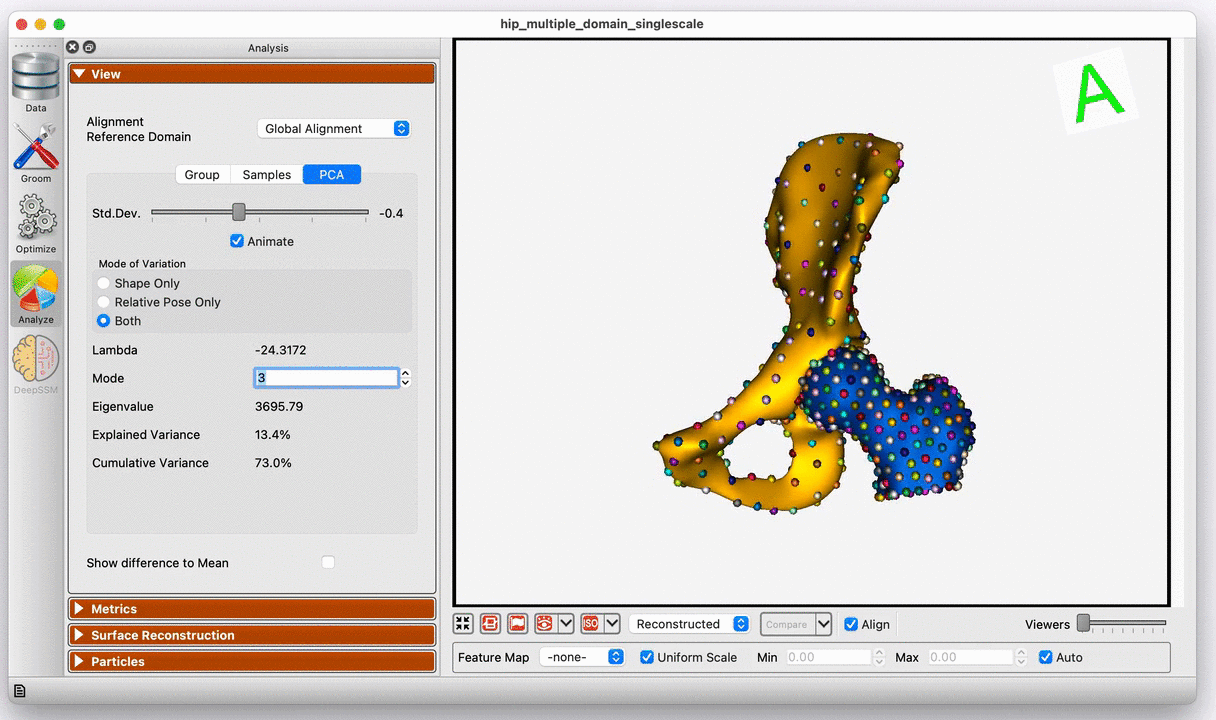
Modes of variation of the hip joint by aligning w.r.t the femur
Modes of variation of the hip joint by aligning w.r.t the pelvis