ShapeWorks Command
ShapeWorks was a conglomeration of independent executables for grooming and optimization with a GUI (ShapeWorks Studio) for analysis and visualization. This design is highly inflexible, task specific, and poorly documented, and Studio duplicated a significant portion of their functionality.
We have made significant efforts in organizing the codebase based on functionalities, implementing them as libraries rather than executables to provide a common backbone to command-line and GUI-based tools, and syncing ShapeWorks Studio to use the same underlying libraries.
To retain command line usage, we have created a single shapeworks command with subcommands exposing this functionality along with greater flexibility and interactive --help for each subcommand.
This consolidation makes the framework more powerful and flexible. It also enables ShapeWorks functionality to be used as libraries linked to new applications.
All the executables used for the segmentation-driven grooming have been consolidated, documented, tested against the original command line tools, and functionally debugged
Comprehensive unit testing is implemented and executed as part of automatic validation run with each addition to the code. This also serves as independent examples of its use
Example: ResampleVolumesToBeIsotropic
Old command-line: ResampleVolumesToBeIsotropic
./ResampleVolumesToBeIsotropic --inFilename <input-file> --outFilename <output-file>
--isoSpacing <voxel-spacing>
[--isBinaryImage] [--isCenterImageOn]
Disadvantages of the old command-line tool:
- Cannot be used by other classes or other APIs or other functions
- Not adaptable (need to edit script files to customize it)
- Each command needs to be given input and output paths
- Creates IO bottlenecks
- Fixed parameters cannot be changed (e.g., num iterations for binarization)
- All logic is buried behind a single command line tool
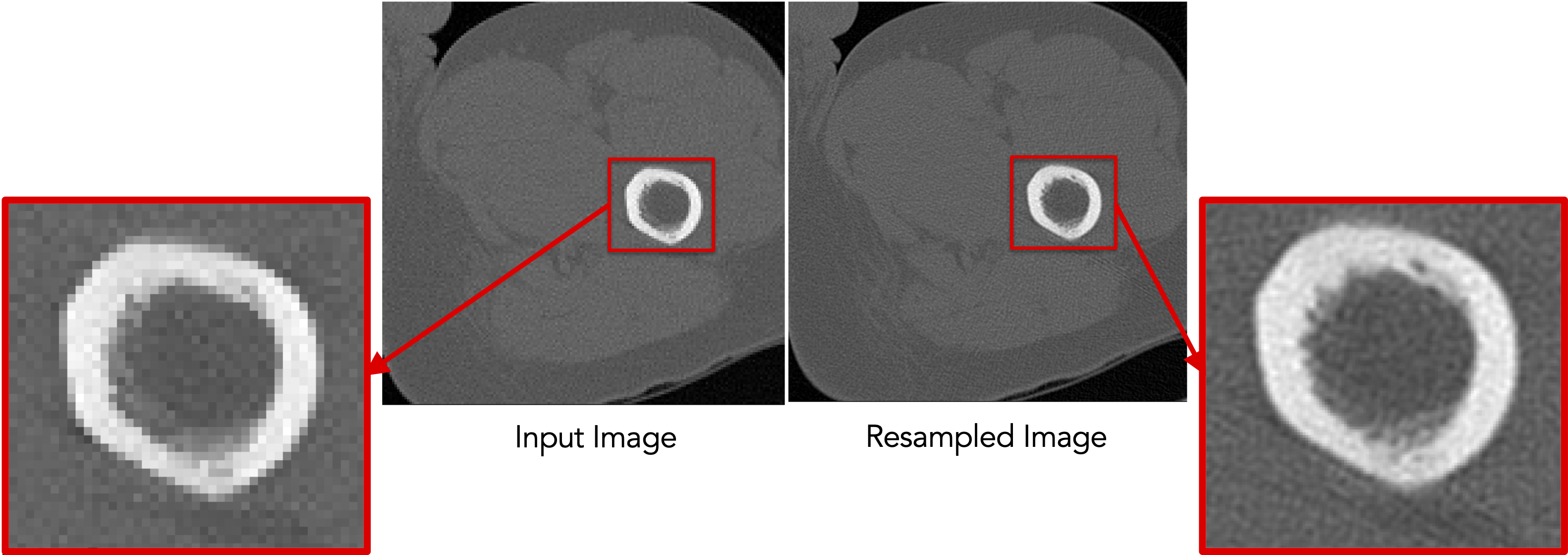
Resampling images
Old command-line: ResampleVolumesToBeIsotropic (for images)
./ResampleVolumesToBeIsotropic --inFilename <input-file> --outFilename <output-file>
--isoSpacing <voxel-spacing>
--isCenterImageOn
New command-line: isoresample (for images)
shapeworks readimage --name <input-file> recenter
isoresample --isospacing <voxel-spacing>
writeimage --name <output-file>
C++ (without chaining): isoresample (for images)
Image img(<input-file>);
img.recenter();
img.isoresample(<voxel-spacing>);
img.write(<output-file>);
C++ (with chaining): isoresample (for images)
Image img(<input-file>).recenter().isoresample(<voxel-spacing>).write(<output-file>);
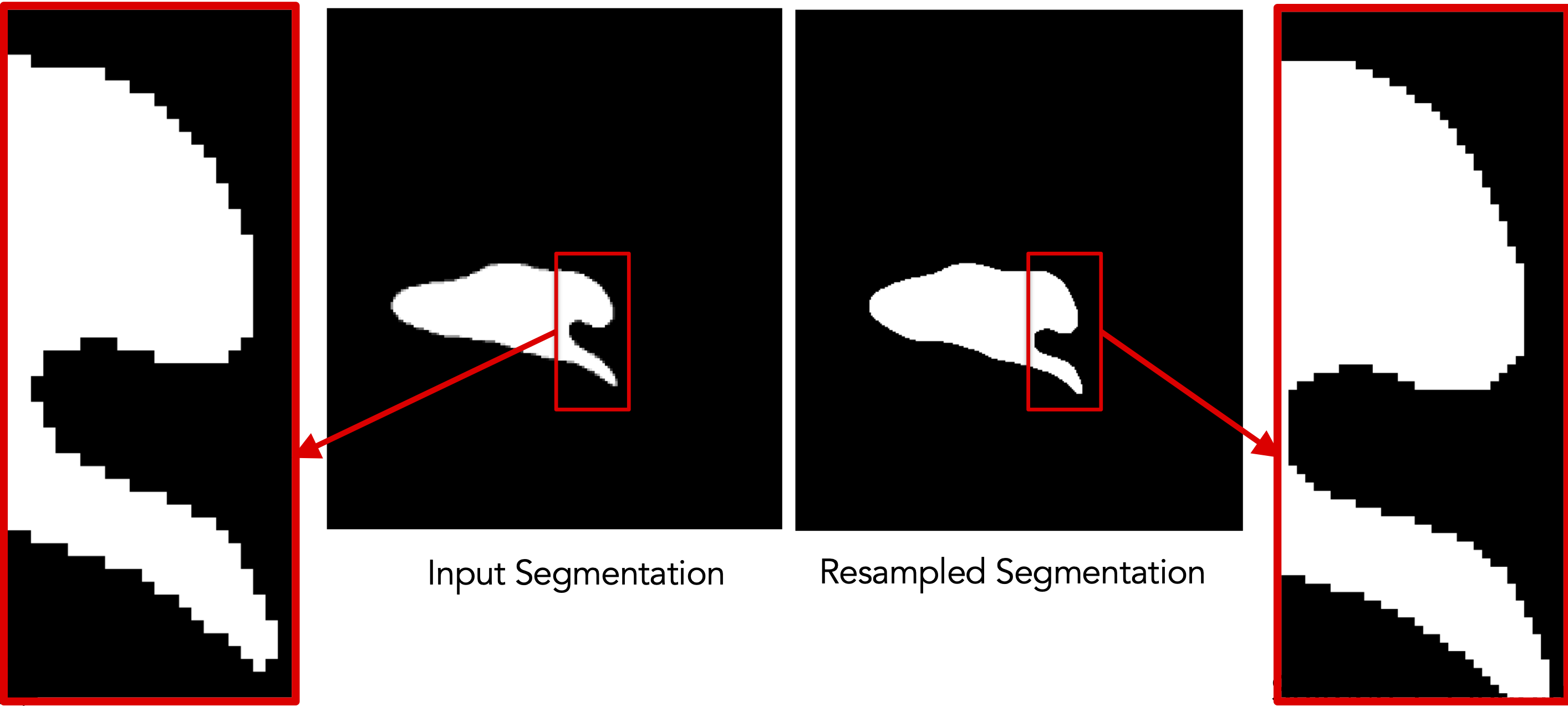
Resampling segmentations
Old command-line: ResampleVolumesToBeIsotropic (for segmentations)
./ResampleVolumesToBeIsotropic --inFilename <input-file> --outFilename <output-file>
--isoSpacing <voxel-spacing>
--isBinaryImage --isCenterImageOn
The old executable’s functionalities are broken down further to make it more modular:
- Antialias using
shapeworks antialias - Recenter using
shapeworks recenter - Binarize using
shapeworks binarize
Advantages for the new shapeworks API:
- Promotes user’s understanding of the underlying functionality (more transparency and equivalent simplicity)
- Allows the user to choose the set of commands to be run
- User can know what parameters are considered to perform each command
- User can modify parameter values each step of the way
- User can save/visualize intermediate outputs for troubleshooting
New command-line: isoresample (for segmentations)
shapeworks readimage --name <input-file>
recenter antialias --iterations <num-iter>
isoresample --isospacing <voxel-spacing> binarize
writeimage --name <output-file>
C++ (without chaining): isoresample (for segmentations)
Image img(<input-file>);
img.recenter();
img.antialias(<num-iter>);
img.isoresample(<voxel-spacing>);
img.binarize();
img.write(<output-file>);
C++ (with chaining): isoresample (for images)
Image img(<input-file>).recenter().antialias(<num-iter>).isoresample(<voxel-spacing>).binarize().write(<output-file>);