Lumps: Shape Model directly from Mesh
What is the Use Case?
The lumps use case demonstrates a minimal example of running ShapeWorks directly on a mesh using a synthetic dataset. The shapes in this dataset are spheres with two lumps or nodes that vary in size. The use case demonstrates that the ShapeWorks workflow results in a correct shape model- i.e., only the position of particles on the lumps vary; the rest are constant across the shape population.
Grooming Steps
This is a synthetic dataset that is already in alignment and does not require grooming.
Here are some examples of the meshes:
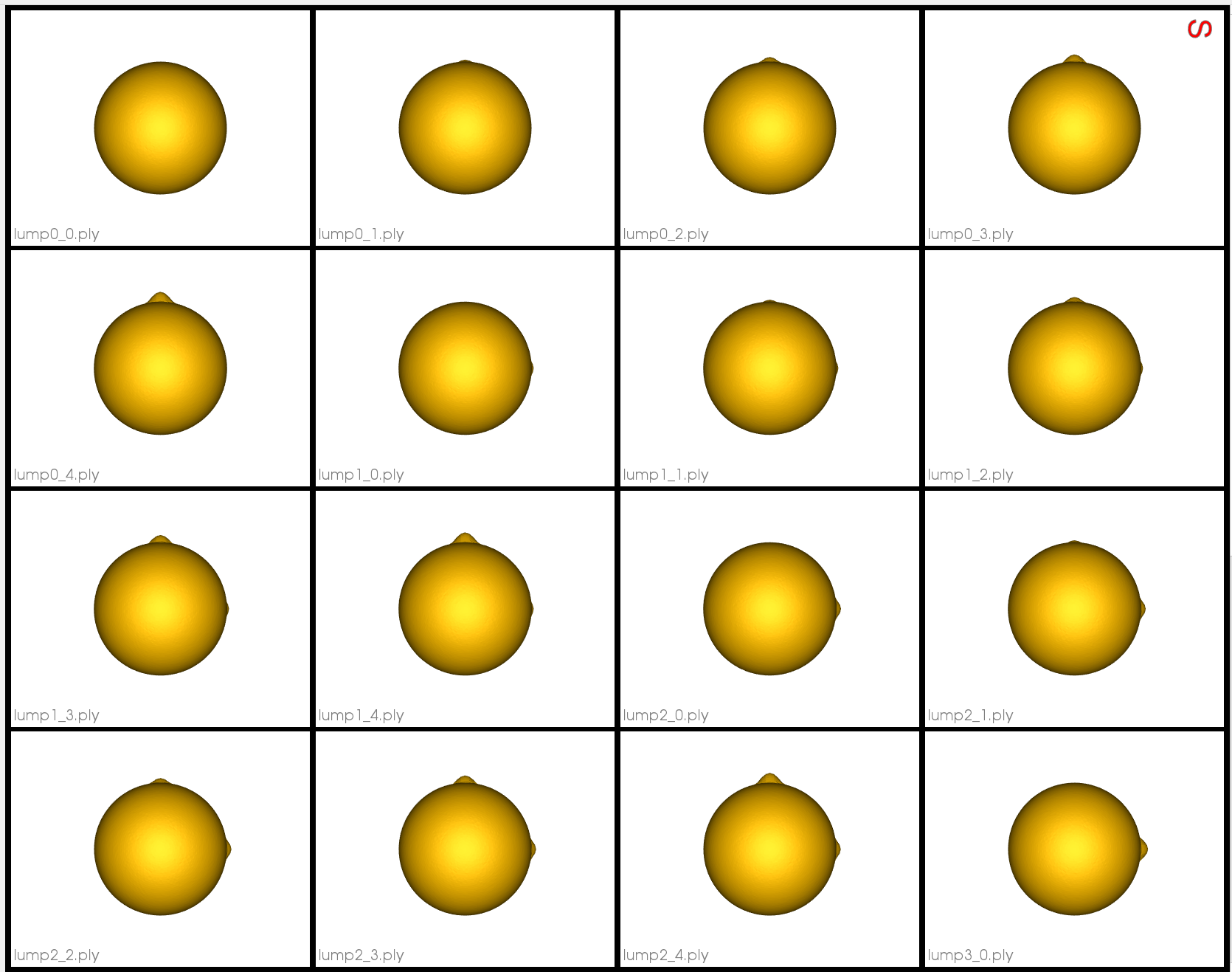
Relevant Arguments
--use_subsample --num_subsample --use_single_scale --tiny_test
Optimization Parameters
The python code for the use case calls the optimize command of ShapeWorks, which requires that the optimization parameters are specified in a python dictionary. Please refer to Parameter Dictionary in Python for more details.
Below are the default optimization parameters for this use case.
{
"number_of_particles": 512,
"use_normals": 0,
"normal_weight": 10.0,
"checkpointing_interval": 100,
"keep_checkpoints": 0,
"iterations_per_split": 2000,
"optimization_iterations": 500,
"starting_regularization": 10,
"ending_regularization": 1,ss
"recompute_regularization_interval": 1,
"domains_per_shape": 1,
"domain_type": "mesh",
"relative_weighting": 10,
"initial_relative_weighting": 1,
"procrustes_interval": 0,
"procrustes_scaling": 0,
"save_init_splits": 0,
"verbosity": 1
}
Analyzing Shape Model
Here is the mean shape of the optimized shape mode using single-scale optimization. Note the two tiny lumps at the top, and towards the right.
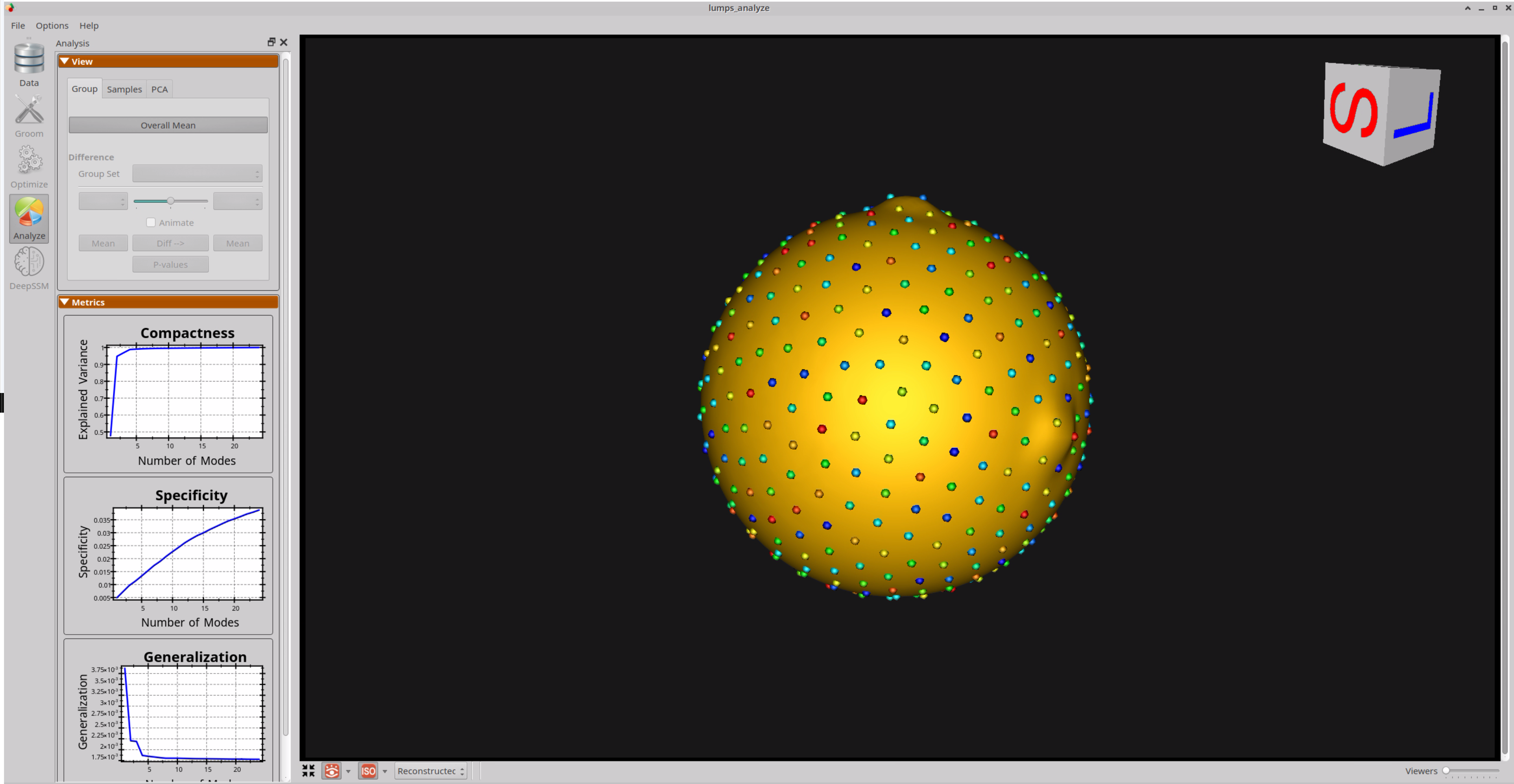
Here are lumps samples with their optimized correspondences.

Here is a video showing the shape modes of variation (computed using principal component analysis - PCA) of the lumps dataset using single-scale optimization.
Note that the particles which do not lie on the lumps remain stationary. The shape model correctly caputures the modes of variation.