Femur Shape Model Directly from Images
What is the Use Case?
The deep_ssm use case demonstrates how to get shape models from unsegmented images using deep learning on the femur data. This includes performing data augmentation as well as building, training and testing a DeepSSM model. For a detailed description of these processes, please see Data Augmentation for Deep Learning and SSMs Directly from Images. The image and shape data used for training and testing results from running the femur use case. Note running this use case does not require running the femur use case, the required data will automatically be downloaded.
On CUDA
This use case uses Pytorch and requires a GPU to run in a timely manner. When you source install_shapeworks.sh, it detects if you have a GPU and installs the version of Pytorch compatible with your version of CUDA.
Note we only support the three most recent versions of CUDA. If your GPU requires an older CUDA version, you will need to update the Pytorch install in your shapeworks conda environment to the correct CUDA version. For more information on doing so, see pytorch.org.
To do a quick check to see if Pytorch is running on your GPU, you can run the use case with the --tiny-test tag. This will quickly run the use case on a few examples and print an error if it is not running on the GPU.
Relevant Arguments
Use Case Pipeline
The use case pipeline includes data augmentation and creating and training a DeepSSM network using ShapeWorks Python packages. For a full explanation of these packages see Using the Data Augmentation Python Package and Using the DeepSSM Python Package.
Step 1: Getting the original data
The femur data is downloaded from the ShapeWorks Data Portal. This use case uses the original unsegmented images and the corresponding .particles files in the shape_models folder downloaded with the femur dataset. Of the 50 examples in the femur dataset, 40 are used to create training and validation sets, while the remaining 10 are saved for a test set (i.e., held out data).
Step 2: Running data augmentation
Data augmentation is run using the images and particle files allocated for training and validation. 4960 augmented samples are created so that DeepSSM can be trained on 5000 total examples. The data is embedded to 6 dimensions using PCA, preserving 95% of the population variation. A kernel density estimate (KDE) distribution is then fit to the embedded data and used in sampling new shape samples for data augmentation. The real and augmented results are then visualized in a matrix of scatterplots.
The functions relevant to this step are runDataAugmentation and visualizeAugmentation.
Step 3: Creating torch loaders
The images and particle files are reformatted into tensors for training and testing the DeepSSM network. The 5000 original and augmented image/particle pairs are turned into train (80%) and validation (20%) loaders and the images held out for the test set are turned into a test loader. A batch size of 8 is used for optimal GPU capacity. The images in the train, validation, and test sets are downsampled to 75% of their original size to decrease training time.
The functions relevant to this step are getTrainValLoaders and getTestLoader.
Note
If a CUDA memory error occurs when running the use case, the batch size value may need to be decreased.
Step 4: Training DeepSSM
A DeepSSM model is created and trained for 50 epochs. A learning rate of 0.0001 is used, and the validation error is calculated and reported every epoch.
The function relevant to this step is trainDeepSSM.
Step 5: Testing DeepSSM
The trained DeepSSM model is used to predict the PCA scores of the unseen images in the test loader. These scores are then mapped to the particle shape model using the PCA information from data augmentation, and the predicted particles are saved.
The function relevant to this step is testDeepSSM.
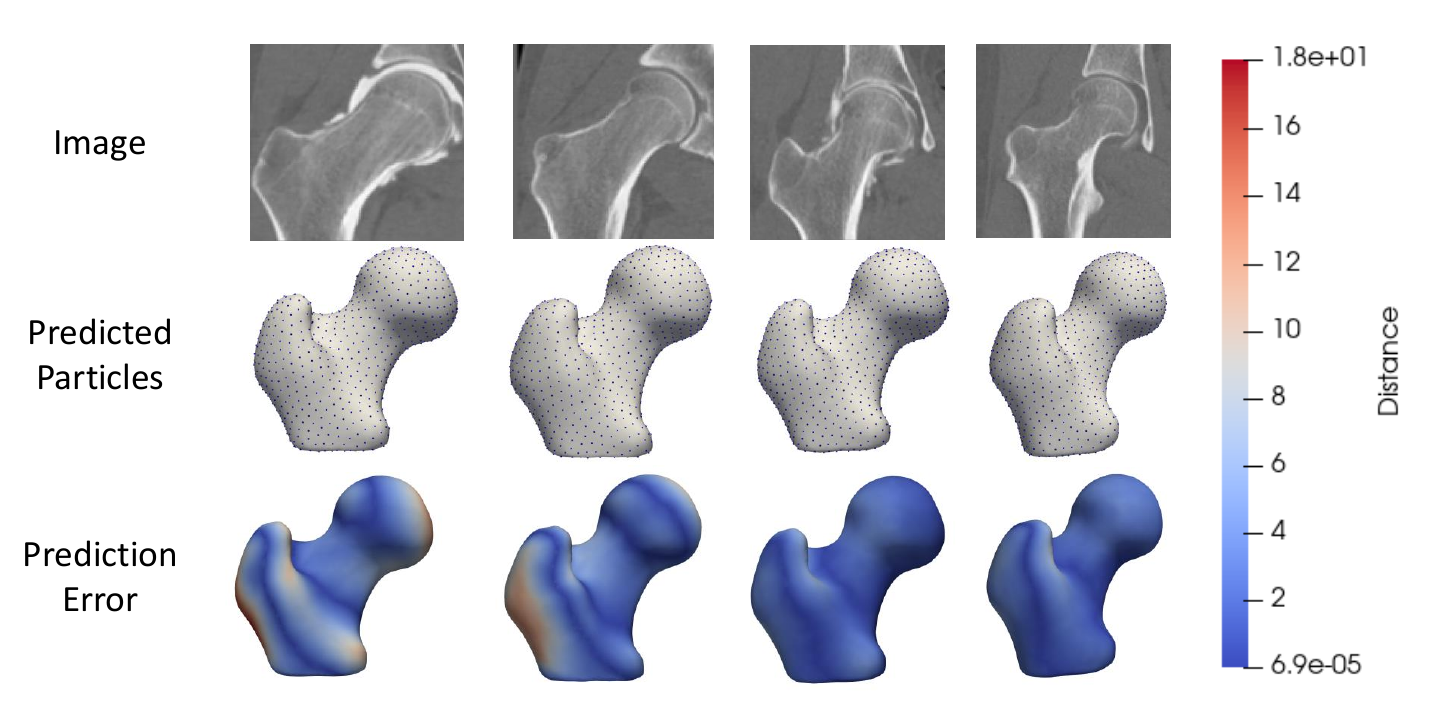
Step 6: Analyze DeepSSM Results
The DeepSSM predictions are analyzed by considering the surface-to-surface distance between the mesh generated from the original segmentation and the mesh generated from the predicted particles. Heat maps of these distances on the meshes are saved from visualizing the results.
The function relevant to this step is analyzeResults.